The FASTQ sequence file format #
The FASTQ sequence file format is widely used for storing biological sequences and their corresponding quality scores. It was originally developed at the Wellcome Trust Sanger Institute to bundle a fasta sequence together with its quality data ( Citation: Cock, Fields & al., 2010 Cock, P., Fields, C., Goto, N., Heuer, M. & Rice, P. (2010). The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic acids research, 38(6). 1767–1771. https://doi.org/10.1093/nar/gkp1137 ) . The format has become the de facto standard for storing the output of high-throughput sequencing instruments.
In FASTQ format, each sequence entry consists of four lines:
- A sequence identifier line beginning with an @ character
- The raw sequence letters using the
iupaccode - A separator line beginning with a + character (optionally followed by the same sequence identifier)
- The quality scores encoded in ASCII format
@my_sequence this is my pretty sequence
ACGTTGCAGTACGTTGCAGTACGTTGCAGTACGTTGCAGT
+
CCCCCCC<CcCccbe[`F`accXV<TA\RYU\\ee_e[XZ
The first word after the ‘@’ symbol in the identifier line is the sequence identifier. The rest of the line is a description of the sequence.
The qualities line gives information about the quality scores assigned to each base by the sequencing machine during the sequencing process. It indicates the probability that the base read is incorrectly sequenced.
\[ P(error) = 10^{-\frac{Q}{10}} \]Sequencers typically provide quality scores in the range of \(0\) to \(40\) , which corresponds to a probability of error \(P(Error)\) in the range of \(10^{0} = 1\) to \(10^{-4}\) . The higher the score, the lower the probability of error.
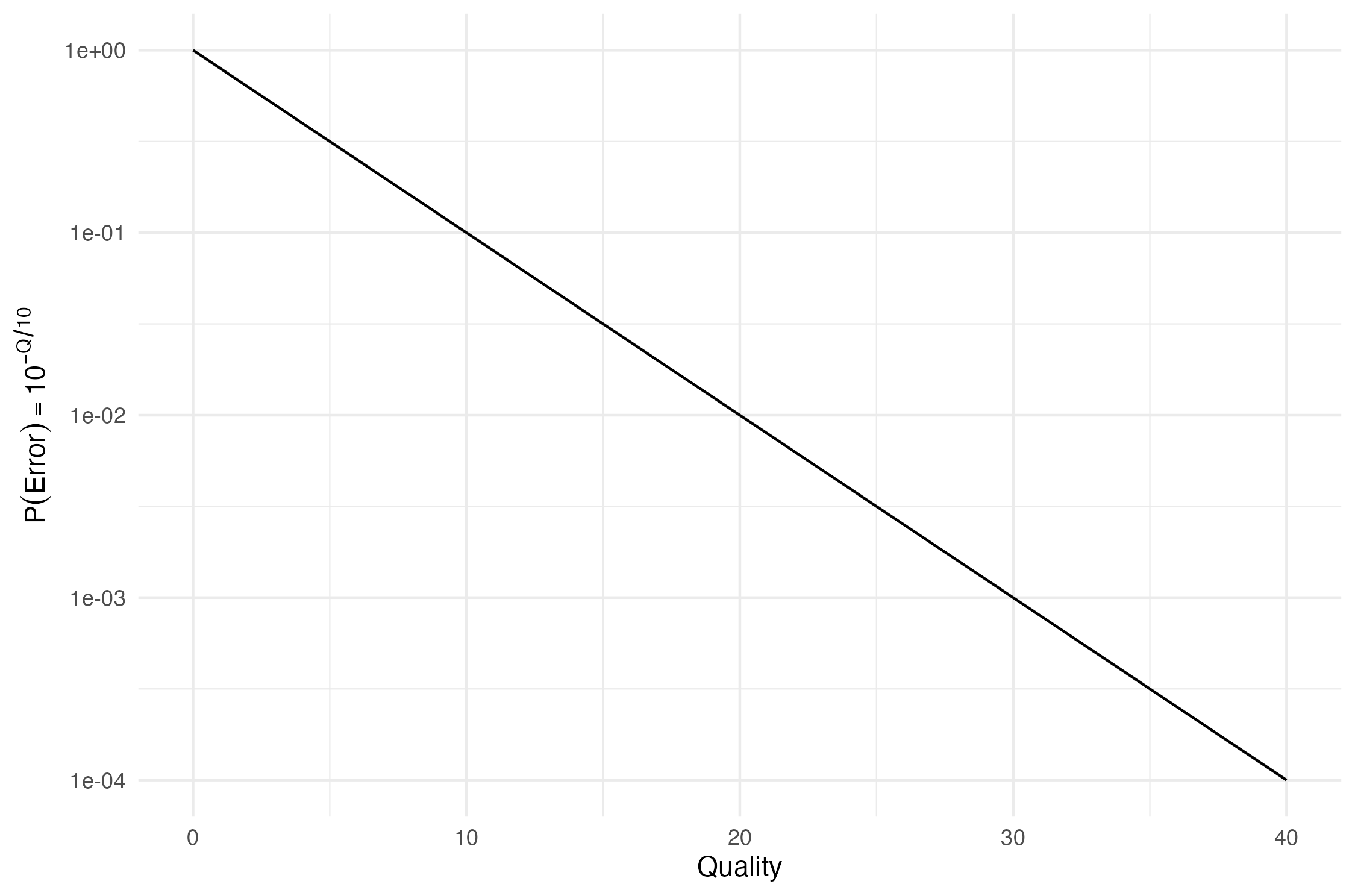
Quality scores and chance of sequencing error
Figure showing the relationship between FASTQ quality scores and error probability
In FASTQ format, the sequence of quality score is encoded as an ASCII string where each score is mapped to an ASCII character. The quality score \(0\)
is encoded as the character !. The quality score \(40\)
is encoded as the character I (uppercase i).
The OBITools extend this format by adding structured data to the identifier line. In the previous version of the OBITools, the structured data was stored after the sequence identifier in a key=value; format, as shown below. The sequence definition was stored as free text after the last key=value; pair.
@HELIUM_000100422_612GNAAXX:7:108:5640:3823#0/1 ali_length=62; mode=alignment; pairing_mismatches={'(T:26)->(G:13)':62,'(T:34)->(G:18)':48}; score=484; seq_b_single=46; ali_dir=left; score_norm=0.968; seq_a_single=46; seq_ab_match=60; sequence definition here
ccgcctcctttagataccccactatgcttagccctaaacacaagtaattaatataacaaaattgttcgccagagtactaccggcaatagcttaaaactcaaaggacttggcggtgctttatacccttctagaggagcctgttctaaggaggcgg
+
CCCCCCCBCCCCCCCCCCCCCCCCCCCCCCBCCCCCBCCCCCCC<CcCccbe[`F`accXV<TA\RYU\\ee_e[XZ[XEEEEEEEEEE?EEEEEEEEEEDEEEEEEECCCCCCCCCCCCCCCCCCCCCCCACCCCCACCCCCCCCCCCCCCCC
@HELIUM_000100422_612GNAAXX:7:97:14311:19299#0/1 mode=alignment; seq_a_single=46; seq_ab_match=52; score=283; score_norm=0.839; seq_b_single=46; ali_dir=left; ali_length=62; pairing_mismatches={'(A:02)->(G:30)':104,'(A:34)->(G:14)':64,'(C:02)->(A:30)':86,'(C:02)->(T:20)':108,'(C:27)->(G:32)':83,'(C:34)->(G:18)':57,'(T:02)->(G:26)':87,'(T:22)->(G:14)':66,'(T:29)->(G:11)':62,'(T:32)->(G:30)':48}; sequence definition here
ccgcctcctttagataccccactatgcttagccctaaacacaagtaattaatataacaaaattgttcgccagagtactaccggcaatagcttaaaactcaaaggacttggcggtgctttatacccttctagaggagcctgttctaaggaggcgg
ccgcctcctttagataccccactatgcttagccctaaacacaagtaattaatataacaaaattattcgccagagtactaccggcaagagcttaaaactcaaaggacttggcggtgctttatacccttctagaggagcctgttctaaggaggcgg
+
CCCCCCCCCCCCCCCCCCCCCCCBBCCC?BCCCCCBC?CCCC@@;AVA`cWeb_TYC\UIN?IDP8QJMKRPVGLQAFPPc`AbAFB5A4>AAA56A><>8>>F@A><8??@BB+<?;?C@9CCCCCC<CC=CCCCCCCCCBC?CBCCCCC@CC
With OBITools4 a new format has been introduced to store structured data in the identifier line. The key/value annotation pairs are now formatted as a JSON map object. The definition is stored as an additional key/value pair using the key `definition’.
📄 two_sequences_obi4.fastq@HELIUM_000100422_612GNAAXX:7:108:5640:3823#0/1 {"ali_dir":"left","ali_length":62,"mode":"alignment","pairing_mismatches":{"(T:26)->(G:13)":62,"(T:34)->(G:18)":48},"score":484,"score_norm":0.968,"seq_a_single":46,"seq_ab_match":60,"seq_b_single":46,"definition":"sequence definition here"}
ccgcctcctttagataccccactatgcttagccctaaacacaagtaattaatataacaaaattgttcgccagagtactaccggcaatagcttaaaactcaaaggacttggcggtgctttatacccttctagaggagcctgttctaaggaggcgg
+
CCCCCCCBCCCCCCCCCCCCCCCCCCCCCCBCCCCCBCCCCCCC<CcCccbe[`F`accXV<TA\RYU\\ee_e[XZ[XEEEEEEEEEE?EEEEEEEEEEDEEEEEEECCCCCCCCCCCCCCCCCCCCCCCACCCCCACCCCCCCCCCCCCCCC
@HELIUM_000100422_612GNAAXX:7:97:14311:19299#0/1 {"ali_dir":"left","ali_length":62,"mode":"alignment","pairing_mismatches":{"(A:02)->(G:30)":104,"(A:34)->(G:14)":64,"(C:02)->(A:30)":86,"(C:02)->(T:20)":108,"(C:27)->(G:32)":83,"(C:34)->(G:18)":57,"(T:02)->(G:26)":87,"(T:22)->(G:14)":66,"(T:29)->(G:11)":62,"(T:32)->(G:30)":48},"score":283,"score_norm":0.839,"seq_a_single":46,"seq_ab_match":52,"seq_b_single":46,"definition":"sequence definition here"}
ccgcctcctttagataccccactatgcttagccctaaacacaagtaattaatataacaaaattattcgccagagtactaccggcaagagcttaaaactcaaaggacttggcggtgctttatacccttctagaggagcctgttctaaggaggcgg
+
CCCCCCCCCCCCCCCCCCCCCCCBBCCC?BCCCCCBC?CCCC@@;AVA`cWeb_TYC\UIN?IDP8QJMKRPVGLQAFPPc`AbAFB5A4>AAA56A><>8>>F@A><8??@BB+<?;?C@9CCCCCC<CC=CCCCCCCCCBC?CBCCCCC@CC
The obiconvert
command, like all other OBITools4 commands, has two options --output-json-header and --output-OBI-header to choose between the new
JSON format and the old OBITools format. The --output-OBI-header option can be abbreviated to -O. By default, the new
JSON OBITools4 format is used, so only the -O option is really useful if the old format is required for compatibility with another software.
Converting from the new JSON format to the old OBITools format:
obiconvert -O two_sequences_obi4.fastq
@HELIUM_000100422_612GNAAXX:7:108:5640:3823#0/1 ali_length=62; mode=alignment; pairing_mismatches={'(T:26)->(G:13)':62,'(T:34)->(G:18)':48}; score=484; seq_b_single=46; ali_dir=left; score_norm=0.968; seq_a_single=46; seq_ab_match=60; sequence definition here
ccgcctcctttagataccccactatgcttagccctaaacacaagtaattaatataacaaaattgttcgccagagtactaccggcaatagcttaaaactcaaaggacttggcggtgctttatacccttctagaggagcctgttctaaggaggcgg
+
CCCCCCCBCCCCCCCCCCCCCCCCCCCCCCBCCCCCBCCCCCCC<CcCccbe[`F`accXV<TA\RYU\\ee_e[XZ[XEEEEEEEEEE?EEEEEEEEEEDEEEEEEECCCCCCCCCCCCCCCCCCCCCCCACCCCCACCCCCCCCCCCCCCCC
@HELIUM_000100422_612GNAAXX:7:97:14311:19299#0/1 mode=alignment; seq_a_single=46; seq_ab_match=52; score=283; score_norm=0.839; seq_b_single=46; ali_dir=left; ali_length=62; pairing_mismatches={'(A:02)->(G:30)':104,'(A:34)->(G:14)':64,'(C:02)->(A:30)':86,'(C:02)->(T:20)':108,'(C:27)->(G:32)':83,'(C:34)->(G:18)':57,'(T:02)->(G:26)':87,'(T:22)->(G:14)':66,'(T:29)->(G:11)':62,'(T:32)->(G:30)':48}; sequence definition here
ccgcctcctttagataccccactatgcttagccctaaacacaagtaattaatataacaaaattgttcgccagagtactaccggcaatagcttaaaactcaaaggacttggcggtgctttatacccttctagaggagcctgttctaaggaggcgg
ccgcctcctttagataccccactatgcttagccctaaacacaagtaattaatataacaaaattattcgccagagtactaccggcaagagcttaaaactcaaaggacttggcggtgctttatacccttctagaggagcctgttctaaggaggcgg
+
CCCCCCCCCCCCCCCCCCCCCCCBBCCC?BCCCCCBC?CCCC@@;AVA`cWeb_TYC\UIN?IDP8QJMKRPVGLQAFPPc`AbAFB5A4>AAA56A><>8>>F@A><8??@BB+<?;?C@9CCCCCC<CC=CCCCCCCCCBC?CBCCCCC@CC
Converting from the old OBITools format to the new JSON format:
obiconvert two_sequences_obi2.fastq
@HELIUM_000100422_612GNAAXX:7:108:5640:3823#0/1 {"ali_dir":"left","ali_length":62,"mode":"alignment","pairing_mismatches":{"(T:26)->(G:13)":62,"(T:34)->(G:18)":48},"score":484,"score_norm":0.968,"seq_a_single":46,"seq_ab_match":60,"seq_b_single":46,"definition":"sequence definition here"}
ccgcctcctttagataccccactatgcttagccctaaacacaagtaattaatataacaaaattgttcgccagagtactaccggcaatagcttaaaactcaaaggacttggcggtgctttatacccttctagaggagcctgttctaaggaggcgg
+
CCCCCCCBCCCCCCCCCCCCCCCCCCCCCCBCCCCCBCCCCCCC<CcCccbe[`F`accXV<TA\RYU\\ee_e[XZ[XEEEEEEEEEE?EEEEEEEEEEDEEEEEEECCCCCCCCCCCCCCCCCCCCCCCACCCCCACCCCCCCCCCCCCCCC
@HELIUM_000100422_612GNAAXX:7:97:14311:19299#0/1 {"ali_dir":"left","ali_length":62,"mode":"alignment","pairing_mismatches":{"(A:02)->(G:30)":104,"(A:34)->(G:14)":64,"(C:02)->(A:30)":86,"(C:02)->(T:20)":108,"(C:27)->(G:32)":83,"(C:34)->(G:18)":57,"(T:02)->(G:26)":87,"(T:22)->(G:14)":66,"(T:29)->(G:11)":62,"(T:32)->(G:30)":48},"score":283,"score_norm":0.839,"seq_a_single":46,"seq_ab_match":52,"seq_b_single":46,"definition":"sequence definition here"}
ccgcctcctttagataccccactatgcttagccctaaacacaagtaattaatataacaaaattattcgccagagtactaccggcaagagcttaaaactcaaaggacttggcggtgctttatacccttctagaggagcctgttctaaggaggcgg
+
CCCCCCCCCCCCCCCCCCCCCCCBBCCC?BCCCCCBC?CCCC@@;AVA`cWeb_TYC\UIN?IDP8QJMKRPVGLQAFPPc`AbAFB5A4>AAA56A><>8>>F@A><8??@BB+<?;?C@9CCCCCC<CC=CCCCCCCCCBC?CBCCCCC@CC
The actual format of the header is automatically detected when OBITools4 commands read a FASTQ file.
References #
- Cock, Fields, Goto, Heuer & Rice (2010)
- Cock, P., Fields, C., Goto, N., Heuer, M. & Rice, P. (2010). The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic acids research, 38(6). 1767–1771. https://doi.org/10.1093/nar/gkp1137